Comparison of Whole Slide Image-Based Deep Learning Algorithms and Genomic Classifiers for Assessing the Risk of Prostate Cancer Metastasis in Surgically Treated Patients
2024, USCAP
View Details
Comparison of Whole Slide Image-Based Deep Learning Algorithms and Genomic Classifiers for Assessing the Risk of Prostate Cancer Metastasis in Surgically Treated Patients
2024, USCAP
Genomic classifiers improve post-radical prostatectomy (RP) risk stratification compared to conventional clinical-pathologic parameters but are tissue-destructive and expensive. In contrast, artificial intelligence algorithms utilizing diagnostic hematoxylin and eosin (H&E)-stained slides for risk stratification conserve tissue and could be made widely available at point-of-care. We compared the predictive output of a deep learning (DL)-based algorithm applied to H&E-stained whole slide images (WSI) of prostate tumors to commercial genomic classifiers such as Decipher and Prolaris in three diverse RP cohorts with follow-up for metastasis.
Comparison of Pathologist and Deep Learning-Based Prostate Cancer Grading for Prediction of Metastatic Outcomes in Primary Prostate Cancer
2024, USCAP
View Details
Comparison of Pathologist and Deep Learning-Based Prostate Cancer Grading for Prediction of Metastatic Outcomes in Primary Prostate Cancer
2024, USCAP
Gleason grading is the most potent prognostic variable in primary prostate cancer, however, inter-observer variability remains a major issue, particularly where subspeciality-trained pathologists are not available. Artificial intelligence algorithms for prostate cancer grading may improve healthcare equity by ensuring widespread access to standardized, high-quality grading, however, most algorithms have not been tested for performance with respect to oncologic outcomes. Here, we compared deep learning-based Gleason grading for the prediction of metastatic outcomes in three large radical prostatectomy cohorts.
Predicting Prostate Cancer Progression with Deep Learning on Low/Intermediate Risk Needle Core Biopsies
2024, USCAP
View Details
Predicting Prostate Cancer Progression with Deep Learning on Low/Intermediate Risk Needle Core Biopsies
2024, USCAP
Risk Stratification of prostate cancer based on Gleason Grade Group (GG) has been the standard of care to guide disease management. Deep Learning (DL) algorithms trained on whole slide images (WSI) to provide GG have been largely validated by comparison to pathologist-determined GG. However, few studies have assessed the performance of DL grading of low/intermediate risk needle biopsies for predicting relevant clinical endpoints.
Evaluating Leading Commercial and Academic AI-Based Gleason Grading Algorithms “In the Wild”
2024, USCAP
View Details
Evaluating Leading Commercial and Academic AI-Based Gleason Grading Algorithms “In the Wild”
2024, USCAP
Public and commercial Gleason grading algorithms based on artificial intelligence (AI) have demonstrated the ability to improve grading accuracy. The evaluation of commercial algorithms in histopathology happens within clinical trials, while academic algorithms are evaluated on benchmark datasets. Commercial algorithms rarely participate in challenges or report performance on public benchmarks, while the data from clinical trials is not available for the academic community to evaluate algorithms on. This research aims to assess and compare the performance of leading public and commercial Gleason grading algorithms using a crowdsourced real-world dataset.
AIRAQc: Deep Learning for Artefact Identification and Quantification in Digital Pathology
2023, Pathology Visions
DOWNLOAD
View Details
AIRAQc: Deep Learning for Artefact Identification and Quantification in Digital Pathology
2023, Pathology Visions
The quality of a whole slide image (WSI) of a histopathology slide may be impacted by artefacts introduced by fixation, processing, sectioning, and/or scanning. Artefacts in WSIs can render portions or the entirety of histopathology slides unsuitable for various types of analysis, and lead laboratory staff to create additional slides or scans that are less impacted. Quality control in preanalytical workflows to detect these artefacts is time and resource-consuming when performed manually and could lead to increased costs and slowed production timelines. In this study, we present a deep learning system trained to identify and quantify frequently occurring artefacts in WSI of H&E-stained tissues.
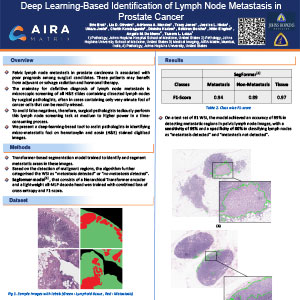
Deep Learning-Based Identification of Lymph Node Metastasis in Prostate Cancer
2023, ECDP
DOWNLOAD
View Details
Deep Learning-Based Identification of Lymph Node Metastasis in Prostate Cancer
2023, ECDP
Pelvic lymph node metastasis in prostate carcinoma is associated with poor prognosis among surgical candidates. These patients may benefit from adjuvant or salvage radiation and hormonal therapy. The mainstay for definitive diagnosis of lymph node metastasis is a microscopic screening of all H&E slides containing dissected lymph nodes by surgical pathologists, often in cases containing only very minute foci of cancer cells that can be readily missed. To avoid false negatives, therefore, surgical pathologists tediously perform this lymph node screening task at medium to higher power in a time-consuming process. We present a deep-learning-based tool to assist pathologists in identifying micro-metastatic foci on hematoxylin and eosin (H&E) stained digitized images.
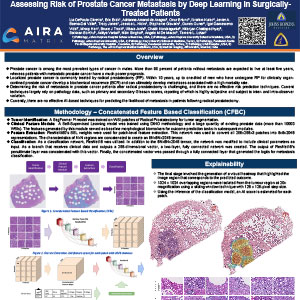
Assessing Risk of Prostate Cancer Metastasis by Deep Learning in Surgically-Treated Patients
2023, ECDP
DOWNLOAD
View Details
Assessing Risk of Prostate Cancer Metastasis by Deep Learning in Surgically-Treated Patients
2023, ECDP
Prostate cancer is among the most prevalent types of cancer in males. More than 90 percent of patients without metastasis are expected to live at least five years, whereas patients with metastatic prostate cancer have a much poorer prognosis. Localized prostate cancer is commonly treated by radical prostatectomy (RP). Within 10 years, up to one-third of men who have undergone RP for clinically organ-confined prostate cancer develop a biochemical recurrence (BCR) and can ultimately develop metastases associated with a high mortality rate. Determining the risk of metastasis in prostate cancer patients after radical prostatectomy is challenging, and there are no effective risk prediction techniques. Current techniques largely rely on pathology data, such as primary and secondary Gleason scores, reporting of which is highly subjective and subject to inter- and intra-observer variation. Currently, there are no effective AI-based techniques for predicting the likelihood of metastasis in patients following radical prostatectomy.
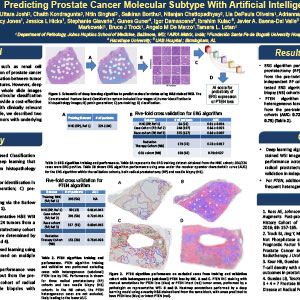
Predicting Prostate Cancer Molecular Subtype with Artificial Intelligence
2023, USCAP
DOWNLOAD
View Details
Predicting Prostate Cancer Molecular Subtype with Artificial Intelligence
2023, USCAP
Unlike other genitourinary cancers, such as renal cell carcinoma, visual microscopic examination of prostate cancer has failed to reveal a reproducible association between tumor molecular subtype and morphologic features. However, deep learning-based algorithms trained on whole slide images (WSI) from large cohorts with known molecular classification may outperform the human eye and provide a cost-effective and rapid method to identify cases with clinically relevant genomic alterations. As proof of principle, we described two such algorithms to identify prostate tumors with underlying ERG fusions and/or PTEN deletion.
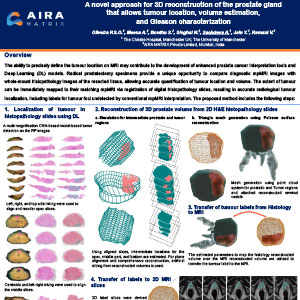
A Novel Approach for 3D Reconstruction of the Prostate Gland that Allows Tumour Location, Volume Estimation, and Gleason Characterization
2022, AUA
DOWNLOAD
View Details
A Novel Approach for 3D Reconstruction of the Prostate Gland that Allows Tumour Location, Volume Estimation, and Gleason Characterization
2022, AUA
The ability to precisely define the tumour location on MRI may contribute to the development of enhanced prostate cancer interpretation tools and Deep Learning (DL) models. Radical prostatectomy specimens provide a unique opportunity to compare diagnostic mpMRI images with whole-mount histopathology images of the resected tissue, allowing accurate quantification of tumour location and volume. The extent of tumour can be immediately mapped to their matching mpMRI via registration of digital histopathology slides, resulting in accurate radiological tumour localization, including labels for tumour foci undetected by conventional mpMRI interpretation.
Deep Learning for Sub-Classification of Gleason Pattern 4 in Prostate Cancer
2022, ECDP
DOWNLOAD
View Details
Deep Learning for Sub-Classification of Gleason Pattern 4 in Prostate Cancer
2022, ECDP
Several studies have provided compelling evidence on the relevance of reporting the percentage of Gleason pattern 4 in a tissue biopsy with a Gleason score of 7. Its significance in risk stratification and therapy planning lets doctors evaluate if a patient is suitable for active surveillance (AS) or definitive therapy, which is why the ISUP has stated that the proportion of Gleason pattern 4 for all tissue samples must be reported in 2019. Furthermore, some studies have suggested that not only the fraction of Gleason pattern 4 present in a sample but also the histologic pattern of Gleason pattern 4 is crucial to record, as it can represent severity and influence prognosis. We created a computational technique that uses Deep Learning to identify and quantify Cribriform and Glomeruloid patterns in whole-mount images (WMI) of radical prostatectomy (RP) specimens.
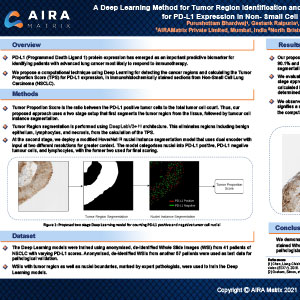
A Deep Learning Method for Tumor Region Identification and Tumor Proportion Score Estimation for PD-L1 Expression in Non-Small Cell Lung Carcinoma
2021, Pathology Visions
DOWNLOAD
View Details
A Deep Learning Method for Tumor Region Identification and Tumor Proportion Score Estimation for PD-L1 Expression in Non-Small Cell Lung Carcinoma
2021, Pathology Visions
The PD-L1 (Programmed Cell Death Ligand 1) protein expression has emerged as a critical biomarker for selecting individuals with advanced lung cancer who are most likely to respond to immune checkpoint inhibitor therapy. The inherent heterogeneous expression of PD-L1 and the availability of multiple PD-L1 assays, detection systems, platforms, and cut-offs have created challenges in ensuring reliable and reproducible reporting which continues to be based on subjective visual assessment by pathologists. Using Deep Learning, we propose a computational technique for recognizing tumor cells in whole slide images (WSI) of immunohistochemically (IHC) stained sections from Non-Small Cell Lung Carcinoma (NSCLC) and accurately computing the Tumour Proportion Score (TPS) for PD-L1 expression.
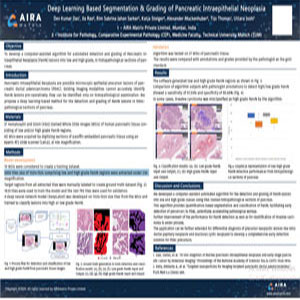
Deep Learning-Based Segmentation & Grading of Pancreatic Intraepithelial Neoplasia
2020, Pathology Visions
DOWNLOAD
View Details
Deep Learning-Based Segmentation & Grading of Pancreatic Intraepithelial Neoplasia
2020, Pathology Visions
To develop a computer-assisted algorithm for automated detection and grading of Pancreatic intraepithelial Neoplasia (PanIN) lesions into low and high grades, in histopathological sections of the pancreas. Pancreatic Intraepithelial Neoplasia are possible microscopic epithelial precursor lesions of pancreatic ductal adenocarcinoma (PDAC). Existing imaging modalities cannot accurately identify PANIn lesions pre-operatively; they can be identified only on histopathological examination. We propose a deep learning-based method for the detection and grading of PanIN lesions in histopathological sections of the pancreas.